HOW CAN WE HELP YOU? Call 1-800-TRY-CHOP
Single Cell Technology Core Services
Let us help you design your next experiment! A well-controlled and balanced study will maximize information and minimize the overall cost. We offer consultations to help you decide the best platform and analysis strategy. Schedule a meeting with core director Dr. Mei Zhang, zhangm5@chop.edu, to learn about all services and technologies available.
Supported Assays
- Single Cell Gene Expression (3′ GEX)
- Single Cell Immune Profiling (VDJ and 5′ GEX)
- Single Cell Assay for Transposase Accessible Chromatin (ATAC)
- Single Cell Multiome (3'GEX + ATAC)
- Single Cell Fixed RNA Profiling
Sample Requirements
Clean single cell or nuclei suspension free of debris and clumping
Cell Viability: >90% ideally, >70% is acceptable
Cell/nuclei concentration:700-1200 cells/ul
Preferred Buffers
1X PBS (calcium and magnesium free) containing 0.04% weight/volume BSA (400 μg/ml).
PBS can be replaced with most common cell culture buffers and media if necessary.
Media should not contain excessive amounts of EDTA (> 0.1mM) or magnesium (> 3mM), as those components will inhibit the reverse transcription reaction.
Any surfactants (Tween-20, etc.) should also be avoided as they may interfere with GEM generation.
For nuclei: include 0.2U/μl RNase Inhibitor (Sigma Protector RNase inhibitor, PN-3335399001 is strongly recommended) in the buffer for gene expression or multiome (gene expression + ATAC) studies.
NanoString's GeoMx Digital Spatial Profiler (DSP)
Supported assays:
- GeoMx Human Whole Transcriptome Atlas: 18,000+ human protein-coding genes
- GeoMx Mouse Whole Transcriptome Atlas: 20,000+ mouse protein-coding genes
- GeoMx Cancer Transcriptome Atlas: 1,800 human genes
Sample Requirements
- Compatible samples: FFPE, Fresh or Fixed Frozen tissue
- For best results, do not use FFPE blocks that are greater than 10 years old
- It is essential to minimize RNases when processing fresh frozen tissues
- Fisherbrand SuperFrost Plus slides or Leica BOND plus slides
- Sectioning: FFPE 5 μm; Frozen 5-10 μm
- Scan area (green area in the picture): 35.3mm x 14.1mm
- Multiple sections can be mounted on the same slide, at least 2-3 mm apart
Vizgen MERSCOPE
Supported assays:
Custom designed gene panels for 140, 300, or 500 genes
Sample Requirements
- Compatible samples: FFPE, Fresh or Fixed Frozen tissue
- Maximum size: 1cm3
- Sectioning: FFPE 4 or 5 μm; Frozen 10 μm
- Mounted on MERSCOPE slides
- RNA quality:
- Frozen tissue: RIN>7 ideal; RIN 5-7: detection efficiency diminishes; RIN<5: do not proceed
- FFPE: DV200>60% yield high quality data; DV200 40-60% yield quality data; DV200<40% yield low quality data
We strongly recommend testing sample quality with the MERSCOPE Sample Verification Kit.
10xGenomics Visium CytAssist
Supported assays:
- Human or mouse whole transcriptome gene expression
- Human or mouse whole transcriptome gene expression and targeted protein expression
Sample Requirements
- Compatible samples: FFPE, Fresh or Fixed Frozen tissue
- Please ensure the desired tissue area will fit within the 6.5 x 6.5 mm or 11 x 11 mm capture area
- There are two capture areas per slide
- Tissue sections should be mounted within the allowable target area on the plain glass slide according to the 10x Genomics Tissue Preparation Guides
- Recommended section thickness:
- FFPE: 3-10 µm
- FF or FxF: 10-20 µm
- RNA quality of the block should be assessed prior to sectioning:
- FFPE: DV200>30%
- FF: RIN>4
- FxF: DV200>50%
10xGenomics Xenium
Supported assays:
- Human or mouse pre-designed panels
- Standalone or add-on custom panels
Sample Requirements
- Compatible samples: FFPE or Fresh Frozen tissue
- Mounted on Xenium slides
- Sample area: 10.45mm x 22.45mm
- Section thickness: FFPE 5 μm; Frozen 10 μm
- Prior to sectioning onto Xenium slides, tissue morphology should be checked for any undesired artifacts
NanoString's GeoMx Digital Spatial Profiler (DSP)
Supported assays
- GeoMx protein assays for nCounter
- GeoMx protein assays for NGS
Sample Requirements
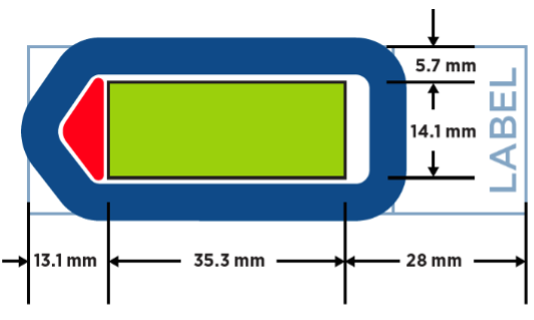
- FFPE, Fresh or Fixed Frozen tissues
- For best results, do not use FFPE blocks that are greater than 10 years old
- Fisherbrand SuperFrost™ Plus slides or Apex BOND® slides
- Sectioning: FFPE 5 μm; Frozen 5-10 μm
- Tissue sections need to be mounted within the scan area (green area in the picture): 35.3mm x 14.1mm
- Multiple sections can be mounted on the same slide, at least 2-3 mm apart
Akoya Phenocycler
Supported assays
- Akoya inventoried antibodies for human fresh frozen and FFPE, and mouse fresh frozen
- Custom conjugated antibodies
Sample Requirements
- FFPE, Fresh or Fixed Frozen tissues
- Coverslips (22mm x 22 mm, Akoyabio) coated with poly-L-lysine
- Tissue sectioning: 5-10 μm (FFPE 4-5 μm, Frozen 8-10 μm); up to 15mm x 15mm
Our experienced bioinformaticians help with experimental design, developing reproducible workflows, analyzing high throughput next-generation sequencing data and spatial profiling data, and supporting manuscript development/publication. We generate visualizations of complex data and assist data uploads to public repositories.